
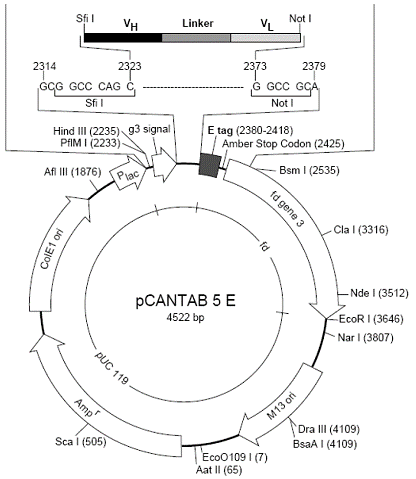
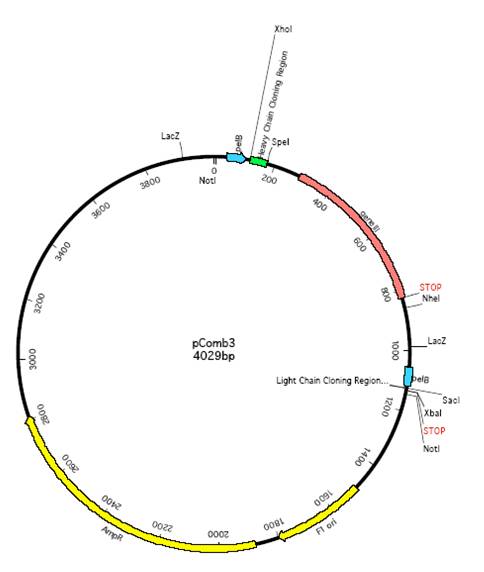
pCANTAB5E, pComb3噬菌体展示系统载体质粒图谱序列抗性
产品名称: pCANTAB5E, pComb3噬菌体展示系统载体质粒图谱序列抗性
英文名称: pCANTAB5E, pComb3噬菌体展示系统载体质粒图谱序列抗性说明书
产品编号: Biovector104802
产品价格: 0
产品产地: Biovector Inc. USA
品牌商标: Biovector
更新时间: null
使用范围: null
pCANTAB5E, pComb3噬菌体展示系统载体质粒图谱序列抗性说明书.辅助噬菌体M13K07, VCSM13.宿主菌TG1,DH5aF,XL1-Blue
Product Information Order ID Name Description Biovector106283 pCANTAB 5E pCANTAB 5E,DNA. 15uL,AmpR.Storage:-20℃ Transformation of plasmid DNA to competent E. Coli cells 1. Thaw competent cells on ice. 20–200µL per tube 2. Add max. 1-3µL of plasmid 3. Mix very gently! 4. Incubate the tubes on ice for 30 min 5. Heat shock the cells for 45 sec to 2 min at 42°C 6. Place the tubes immediately on ice for at least 2 min 7. Add 800µL of SOC medium to each tube 8. Incubate for 1 hour at 37°C and shake vigorously 9. Spin down briefly and remove most supernatants 10. Resuspend cell pellet with the rest SOC medium in the tube by pipetting 11. Plate out the suspension on a LB agar plate containing the appropriate antibiotic. Incubate the plates overnight at 37°C
Product Data Sheet
|
Order ID |
Name |
Description |
|
Biovector108925 |
pComb3 |
pComb3,AmpR. in E.coli. Storage:4℃ |
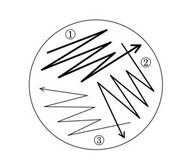 Recovery
Recovery
1. Obtain an LB agar plate with the appropriate antibiotic.
2. Using a sterile pipette tip, touch the bacteria growing within the punctured area of the stab culture. (A sterilized wire loop or sterile toothpick can be used in place of a sterile pipette tip.)
3. Run this tip lightly over a section of the plate, as shown in the figure, to create streak #1.
4. Using another sterile pipette tip, pass through streak #1 and spread the bacteria over a second section of the plate, to create streak #2.
5. Using a third sterile pipette tip, pass through streak #2 and spread the bacteria over the last section of the plate, to create streak #3.
6. Grow overnight in a 37 o C incubator (unless a different growth temperature is indicated on the plasmid datasheet).
7. In the morning, single colonies should be visible. If the bacterial growth is too dense, re-streak onto a new agar plate to obtain single colonies.
Product Information
|
Order ID |
Name |
Description |
|
Biovector-01128569 |
M13KO7 |
M13KO7 helper phage, 100uL. Storage:-20℃ |
Description:
M13KO7 is an M13 derivative which carries the mutation Met40Ile in gII, with the origin of replication from P15A and the kanamycin resistance gene from Tn903 both inserted within the M13 origin of replication (1). M13KO7 is able to replicate in the absence of phagemid DNA. In the presence of a phagemid bearing a wild-type M13 or f1 origin, single-stranded phagemid is packaged preferentially and secreted into the culture medium.This allows easy production of single-stranded phagemid DNA for mutagenesis or sequencing.
Source:
M13KO7 phage supernatant was isolated from infected E. coli ER2738 by a standard procedure.
Concentration:
1 x1011 pfu/ml
Storage Temperature:
-20°C
Use of M13KO7 Helper Phage for isolation of single-stranded phagemid DNA
Protocol![]()
1. Transform phagemid vector into appropriate F strain (CJ236 for Kunkel mutagenesis).
2. Inoculate 50 ml LB (no antibiotic) with a fresh colony, grow at 37°C with vigorous aeration until slightly turbid (<10 Klett, A600 < 0.05). For Kunkel mutagenesis, add uridine to 0.25 µg/ml.
3. Add 50 µl M13KO7 helper phage (final concentration of 1 x 108 pfu/ml), continue vigorous aeration for 60-90 minutes.
4. Add kanamycin to final concentration of 70 µg/ml, grow overnight (14-18 hours) with vigorous aeration.
5. Spin culture at 8,000 rpm for 10 minutes. Transfer supernatant to a new tube and spin again.
6. Pipet the upper 90% of supernatant into a new tube. To this supernatant, add a 0.2 volume of 2.5 M NaCl/20% PEG. Incubate at 4°C for 60 minutes.
7. Recover the phage by centrifugation at 8,000 rpm for 10 minutes. Decant supernatant and spin again briefly.
8. Resuspend the pellet in 1.6 ml TE, transfer to 2 microfuge tubes.
9. Spin in a microfuge for 1 minute to pellet any remaining cells, transfer supernatant to new tubes.
10. Add 200 µl of the 2.5 M NaCl/20% PEG solution to each, let sit at room temperature for 5 minutes, spin in a microfuge for 10 minutes.
11. Decant the supernatant, spin again briefly, remove last traces of supernatant with pipetman.
12.
