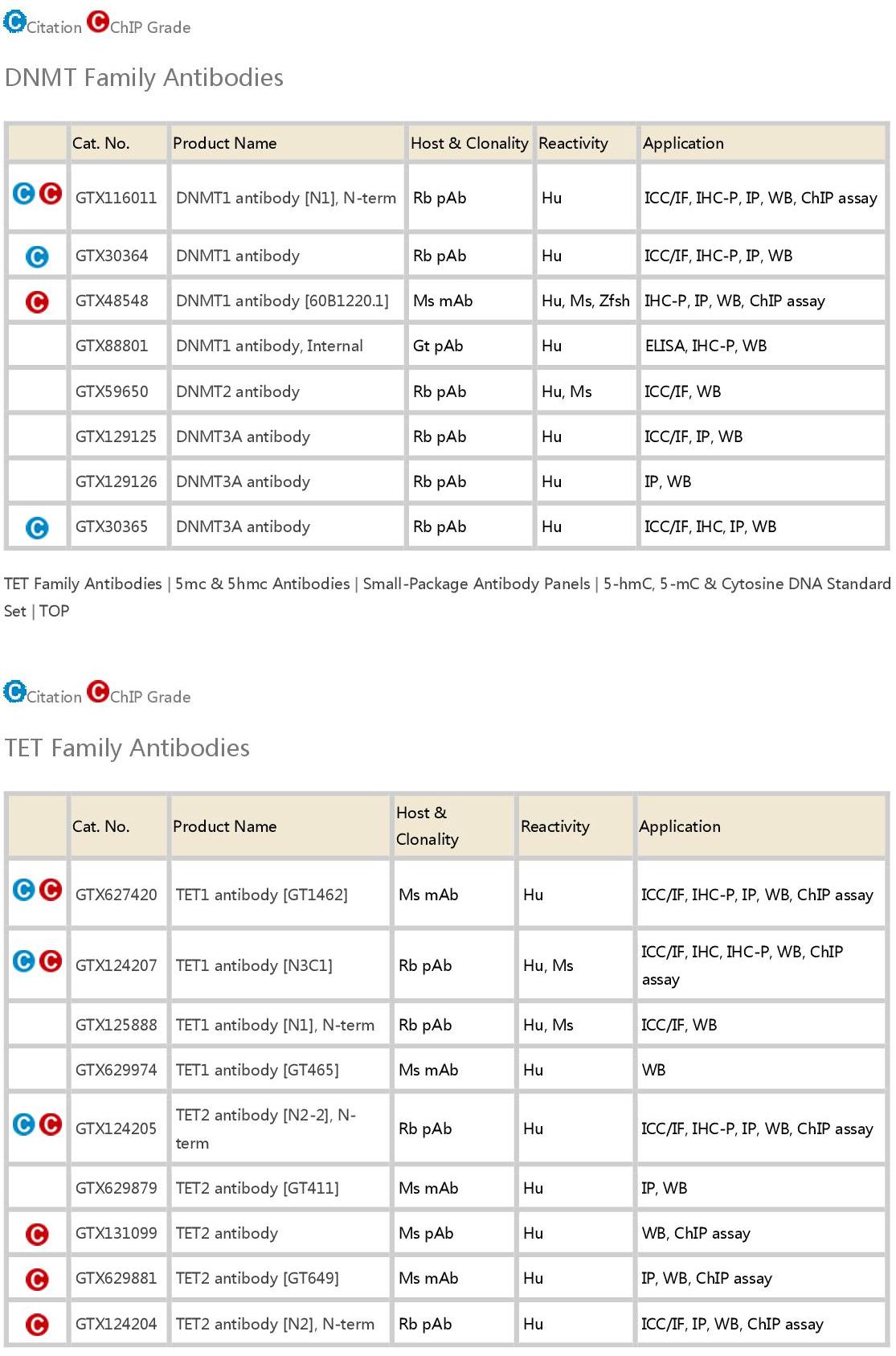
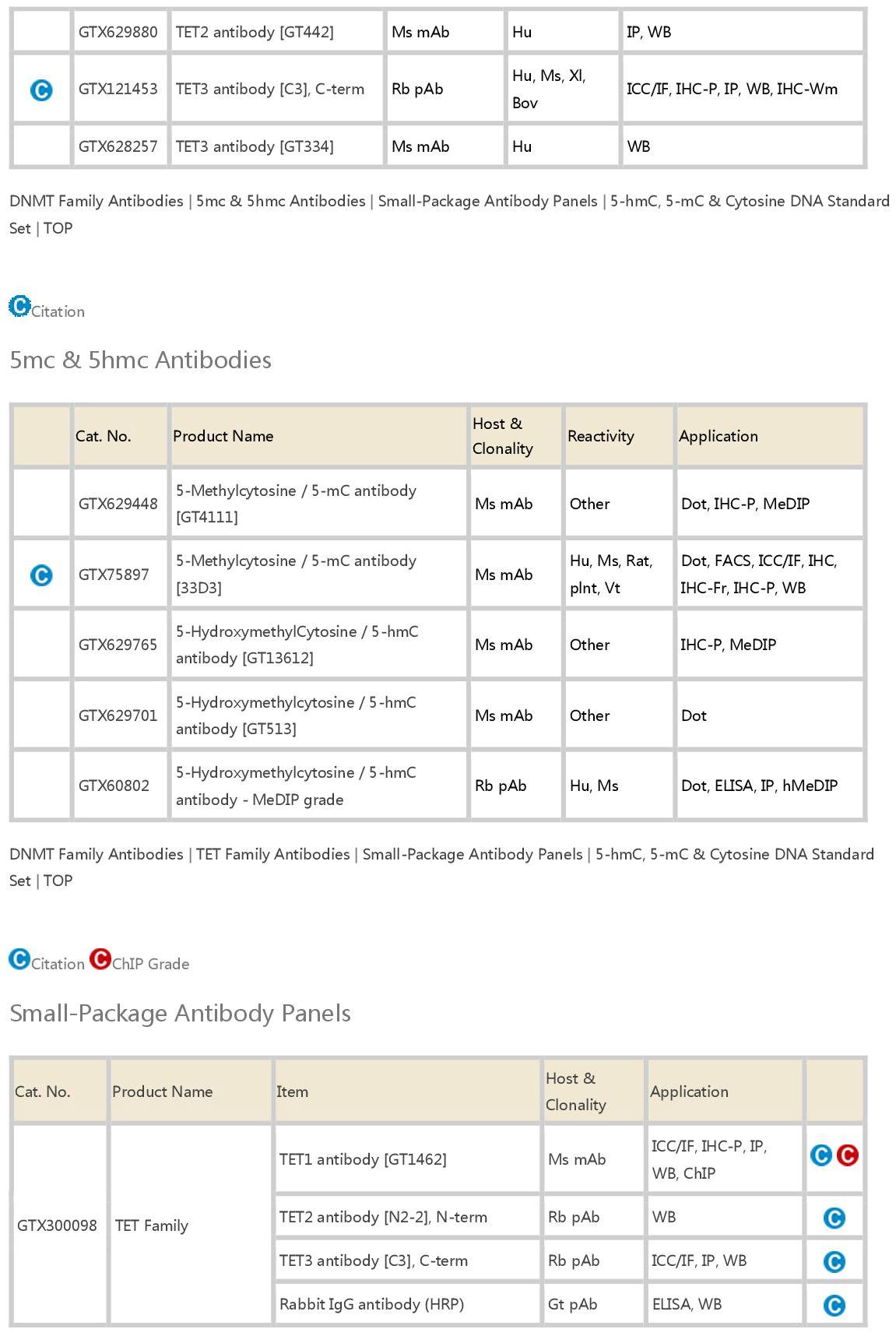
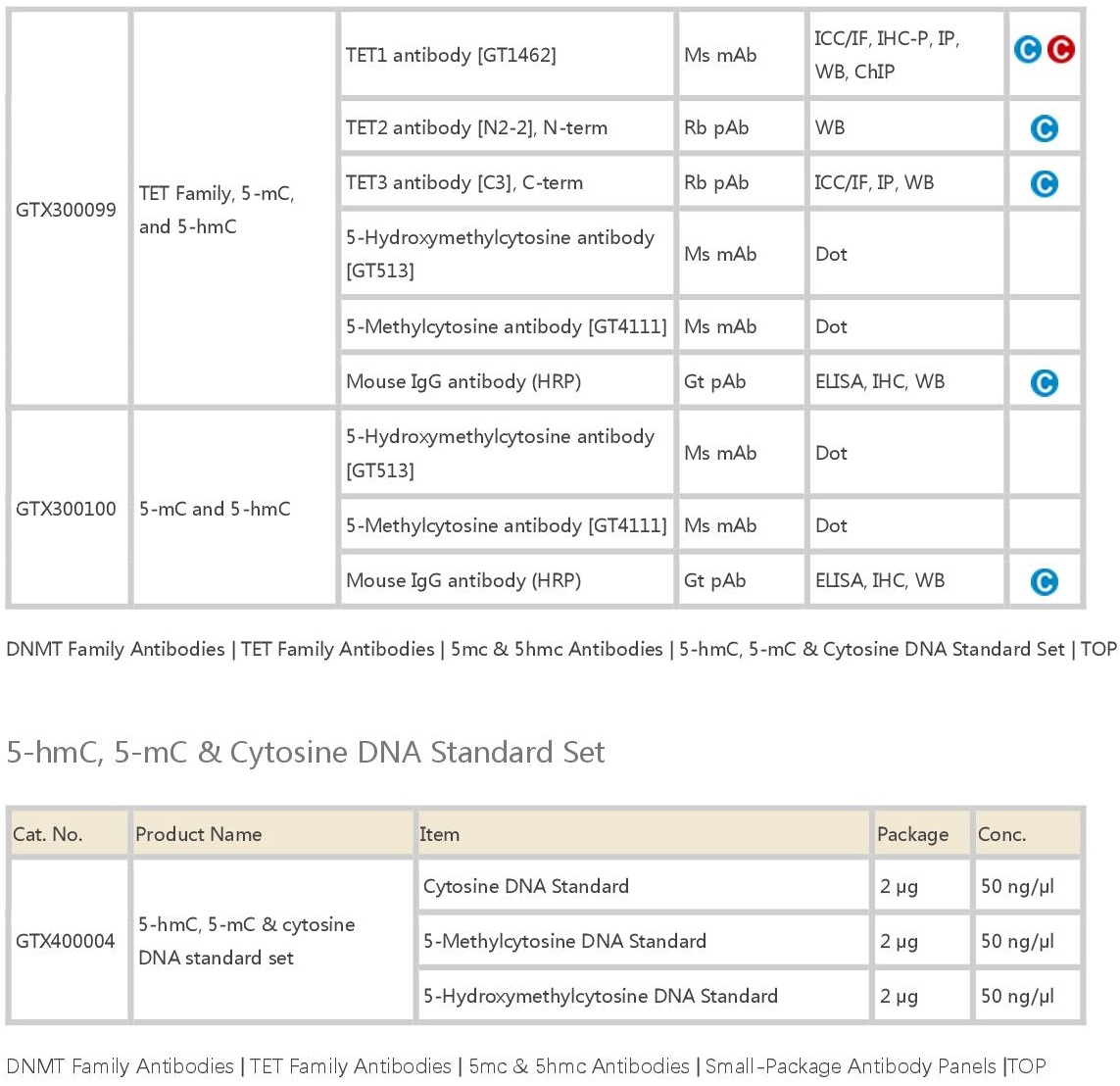
DNA甲基化修饰检测抗体(Antibodies for DNA Methylation)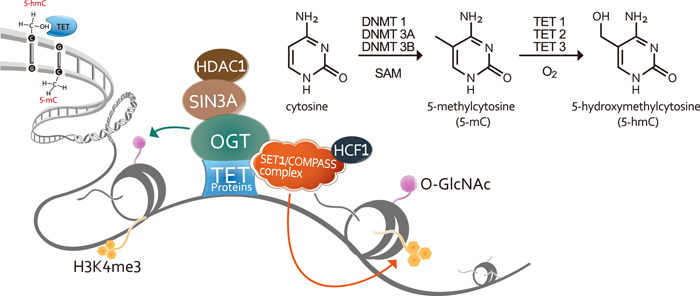
The three mammalian 5-methylcytosine (5-mC) dioxygenases (Ten Eleven Translocation (TET); TET1, TET2, and TET3 in humans) convert 5-mC to 5-hydroxymethylcytosine (5-hmC) and subsequently to 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). This series of modifications is thought to represent one pathway to active DNA demethylation, thereby reshaping the methylated DNA landscape established by DNA methyltransferases (DNMTs). In addition, these marks, particularly 5-hmC, have been shown to be distinct epigenetic signatures in their own right impacting stem cell and cancer cell biology. TET proteins have also been shown to be components of crosstalk mechanisms between DNA and histone modifications that determine gene expression status. Thus, within the span of a few years, the TET proteins have become recognized as central players in gene regulation, pluripotency, cell differentiation, and carcinogenesis. It is clear that researchers have crossed a threshold into an exciting new realm of epigenetics research, with countless discoveries waiting to be made.
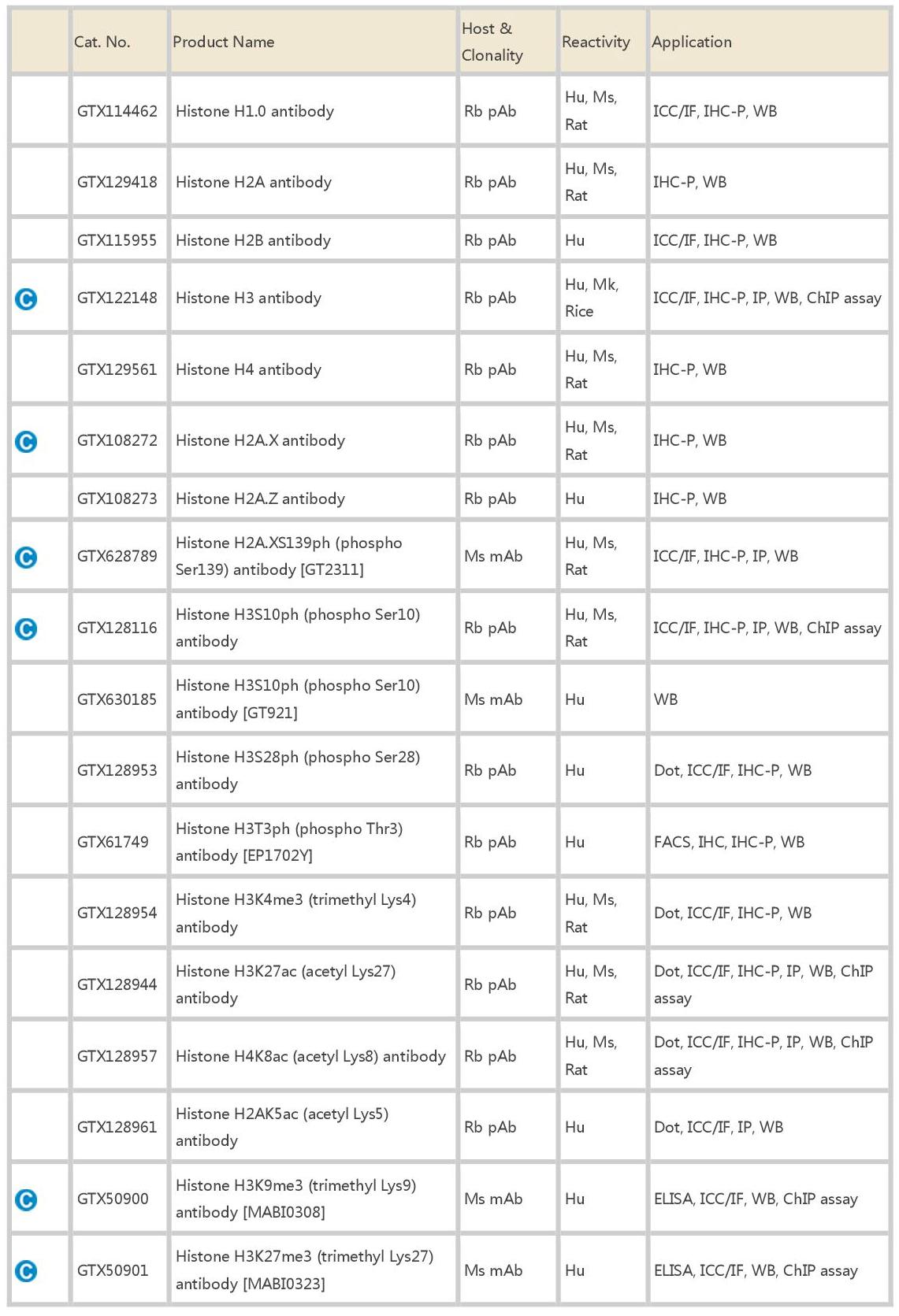
GeneTex is proud to introduce a selection of its antibodies to targets relevant to DNA methylation and demethylation, histone modification, and cell pluripotency. Many of the antibodies are suitable for IP/ChIP application.
Epigenetics Histone Modification
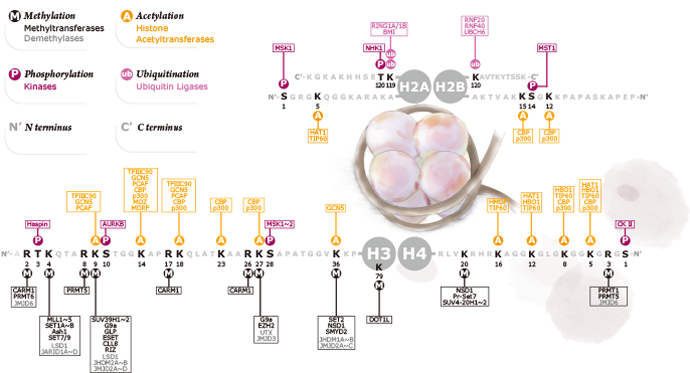
The nucleosome core particle is the fundamental structural unit of the eukaryotic genome. It consists of a histone octamer composed of two H2A-H2B dimers and a H3-H4 tetramer wrapped by ~146 base pairs of DNA. A linker histone (i.e., H1) associates with the nucleosomal dyad as well as with linker DNA on either side of the nucleosome, resulting in the formation of the chromatosome. All the core and linker histones are posttranslationally decorated, with at least 160 total modifications described to date including acetylation, methylation, phosphorylation, propionylation, citrullination, formylation, proline isomerization, butyrylation, ADP ribosylation, ubiquitylation, sumoylation, and the more recently identified glycosylation and crotonylation. These modifications are thought to impact chromatin functions by either altering chromatin packaging or through the recruitment/inhibition of specific chromatin binding factors. Thus, the combined signal from a particular collection of histone marks constitutes a “histone code” that affects gene expression or other chromatin-based functions.
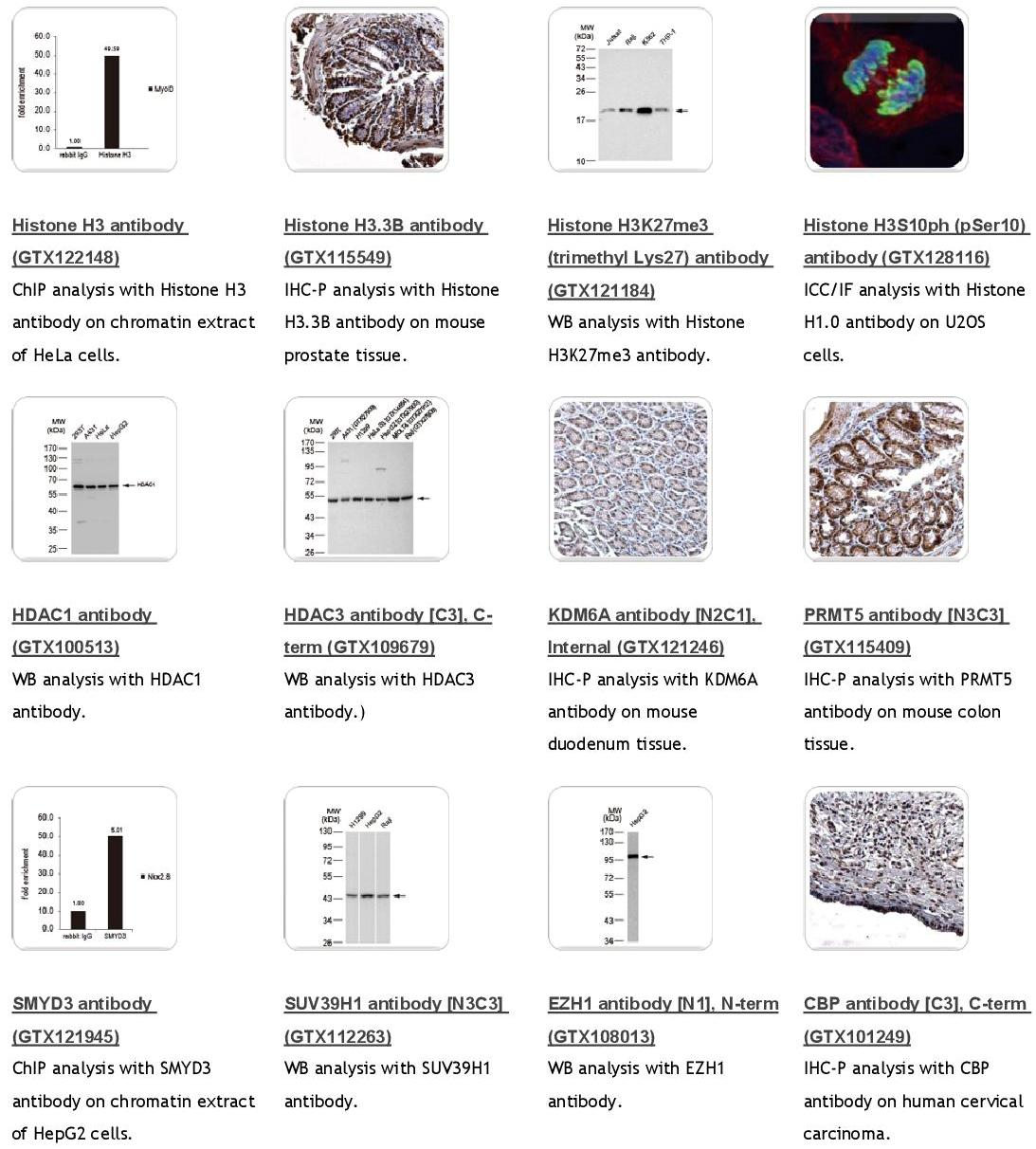


![DNMT1 antibody [N1], N-term (GTX116011)](http://www.genetex.com/fckeditor/uploadfile/tet_001.jpg)


![DNMT3B antibody [52A1018] (GTX50966)](http://www.genetex.com/fckeditor/uploadfile/tet_004.jpg)
![TET1 antibody [GT1462] (GTX627420)](http://www.genetex.com/fckeditor/uploadfile/tet_005.jpg)
![TET2 antibody [N2-2], N-term (GTX124205)](http://www.genetex.com/fckeditor/uploadfile/tet_006.jpg)
![TET3 antibody [C3], C-term (GTX121453)](http://www.genetex.com/fckeditor/uploadfile/tet_007.jpg)
![5-mC antibody [GT4111] (GTX629448)](http://www.genetex.com/fckeditor/uploadfile/tet_008.jpg)